Free and open-source high-throughput virtual screening (HTVS) tools
rDock and RxDock
Fast and versatile open-source programs for docking ligands to proteins and nucleic acids.
- rDock
-
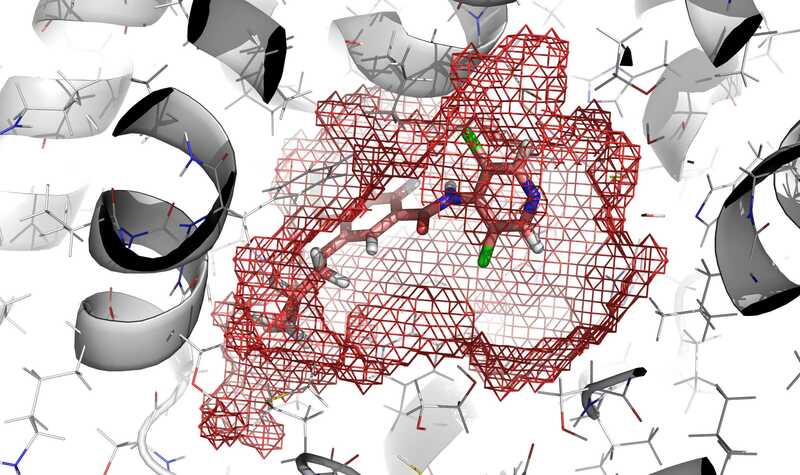
In red mesh, definition of the cavity obtained by execution of
rbcavityprogram.rDock (homepage on SourceForge, former homepage at York Structural Biology Laboratory) is a fast, versatile, and open-source program for docking ligands to proteins and nucleic acids. The source code is available on SourceForge. The functionality is described in the following paper:
Ruiz-Carmona, S., Alvarez-Garcia, D., Foloppe, N., Garmendia-Doval, A. B., Juhos S., et al. (2014) rDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLoS Comput Biol 10(4): e1003571. doi:10.1371/journal.pcbi.1003571
The rDock development has stalled since 2014. - RxDock
-
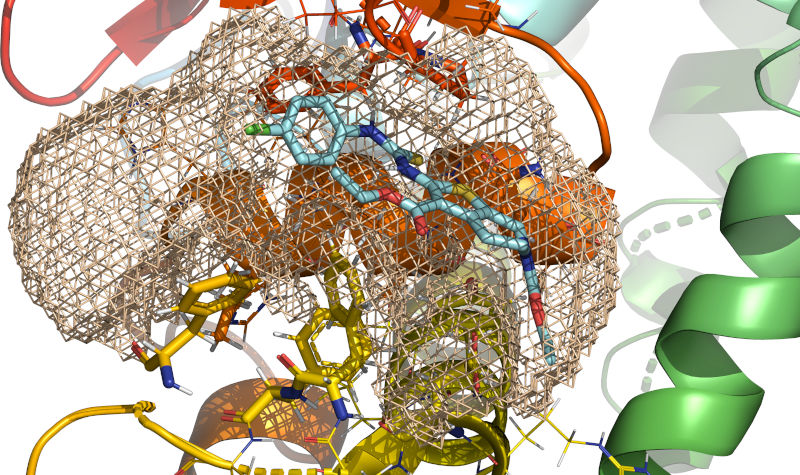
In wheat-colored mesh, definition of the cavity obtained by execution of
rxcmd cavity-searchprogram.RxDock (homepage) is a fork of rDock started in 2019 by RxTx Research with the goal of updating the code for running on modern computer systems (up to and including supercomputers), improving the application programming interfaces, and implementing new functionality. Since its inception, RxDock has gained suport for operating systems other than Linux using recent C++ compilers. The build system, command-line interface, logging, tests, and code organization have been substantially improved. Obsolete linear algebra libraries have been replaced with Eigen. Further improvements are developed on a best-effort basis.
© RxTx Research, 2019 – 2022. First image provided by the rDock project under LGPLv3; second image provided by the RxDock project under LGPLv3.
Neither RxTx Research nor RxTx is in any way affiliated with the rDock project; rDock is supported by University of Barcelona. RxTx Research is hosting and steering the development of the RxDock project; learn more about open source at RxTx.